Overview
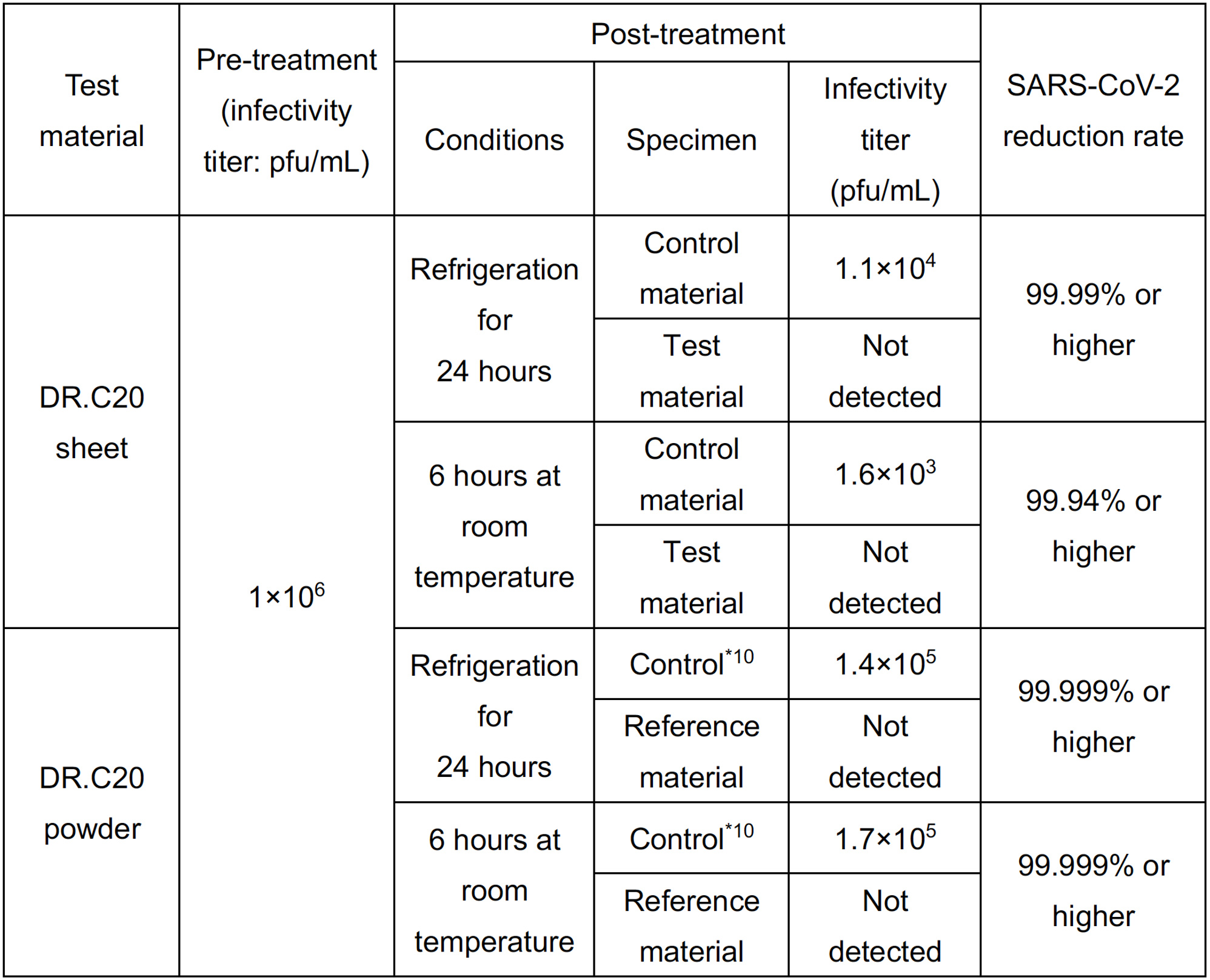
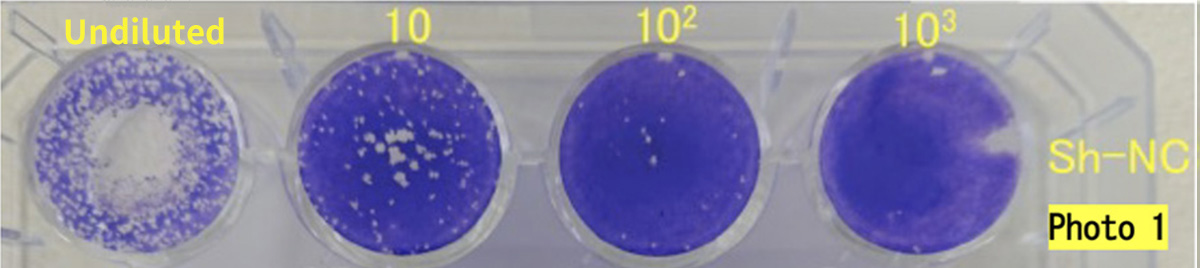
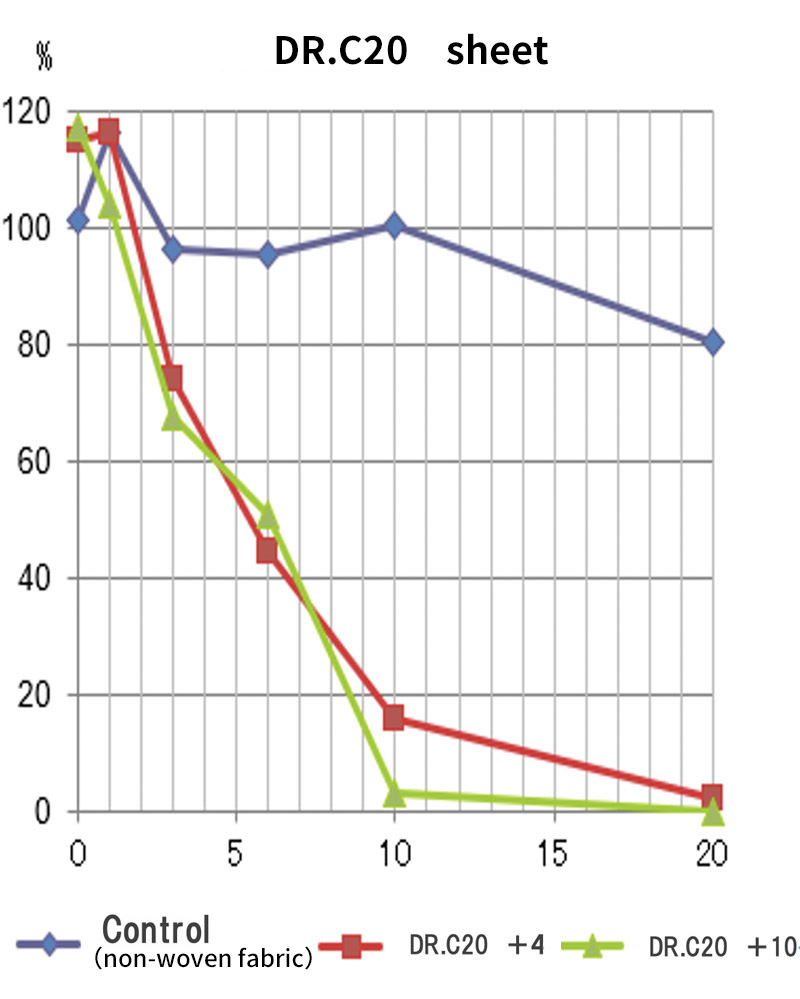
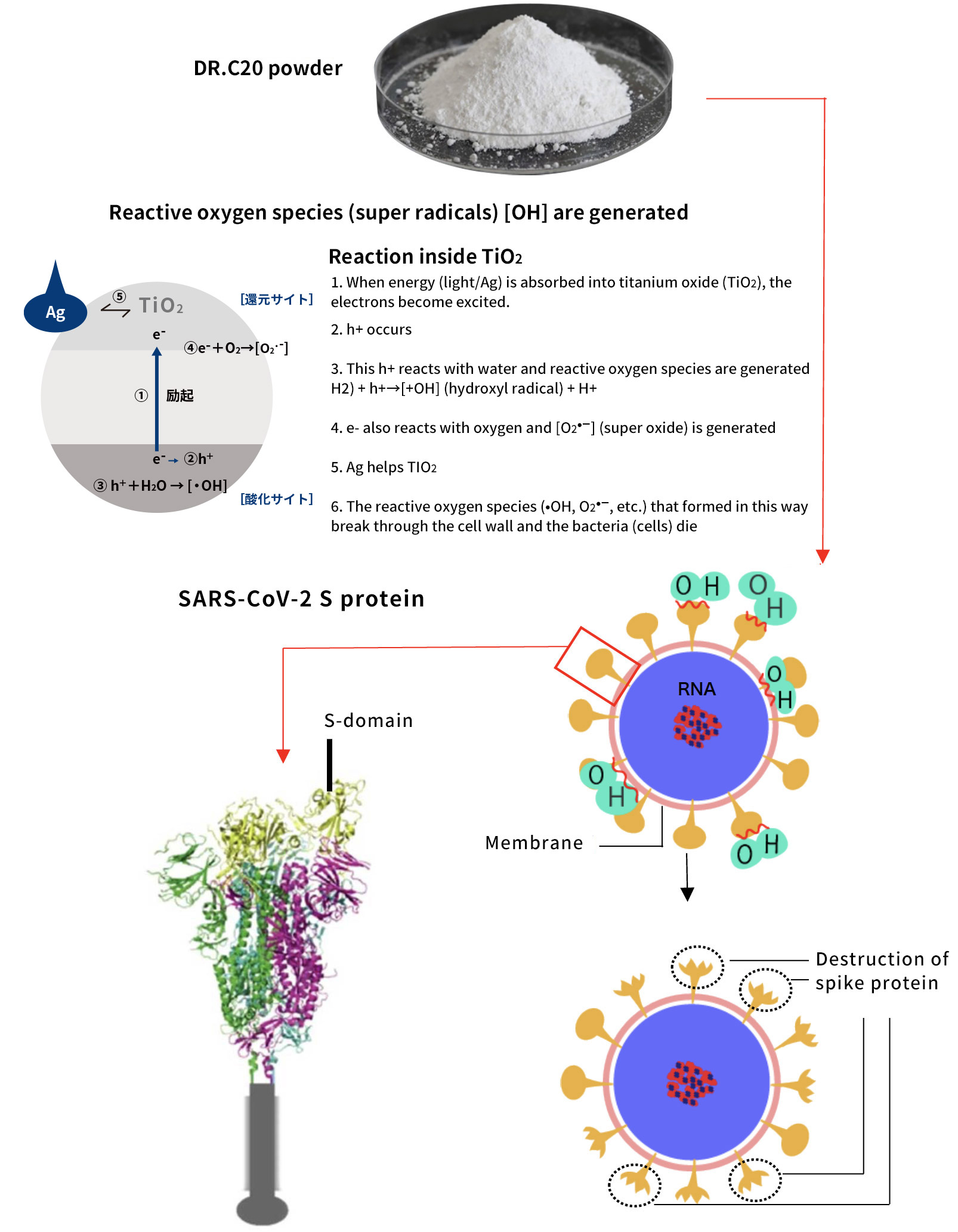
DRC Medicine Ltd. (Representative Director: Narumi Okazaki), in a joint initiative with Professor Jiro Yasuda of the world-renown infectious disease institute, National Research Center for the Control and Prevention of Infectious Diseases Nagasaki University (CCPID) (also a Professor of Emerging Infectious Diseases at the Institute of Tropical Medicine) has confirmed that the SARS-CoV-2 infectivity titer was reduced by 99.99% when a sheet adhered with DR.C20 powder (hereinafter “DR.C20 sheet”) was immersed in SARS-CoV-2 solution and estimates that the infectivity titer will decrease by 99% or more within 5 minutes
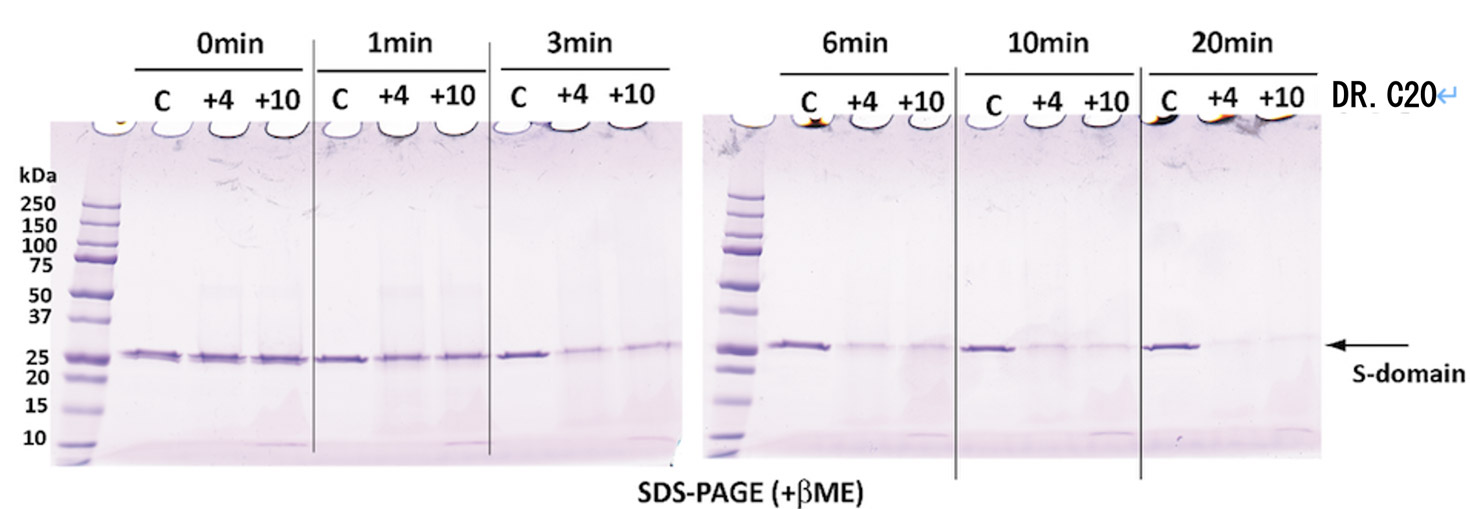
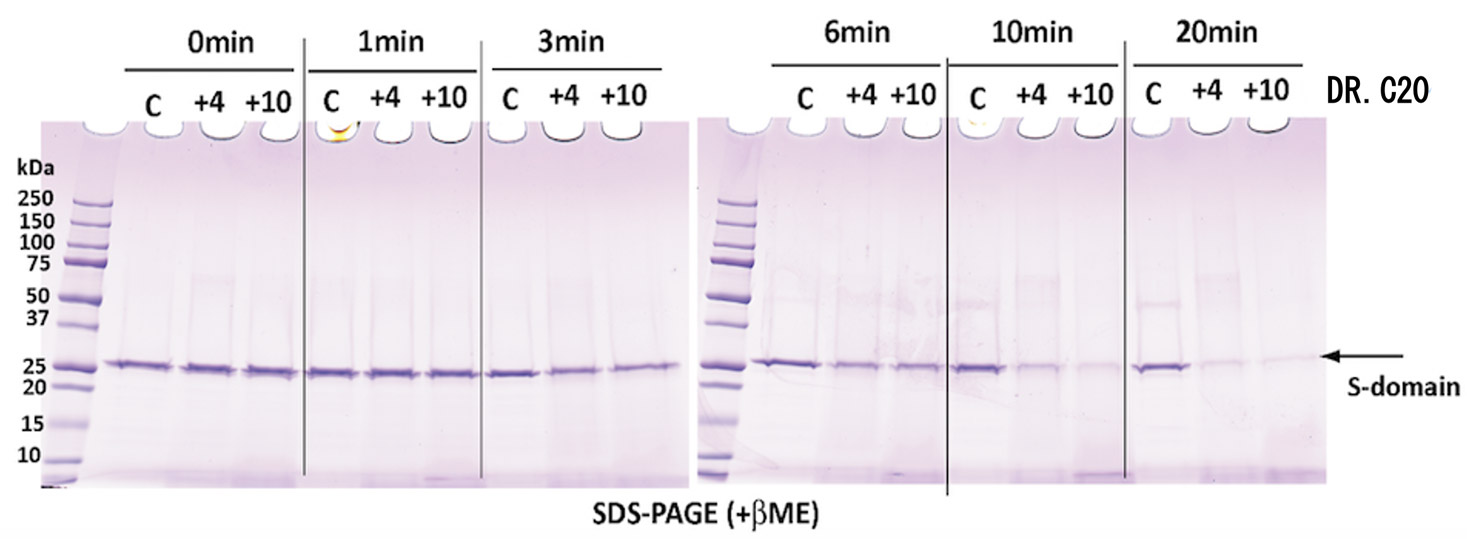
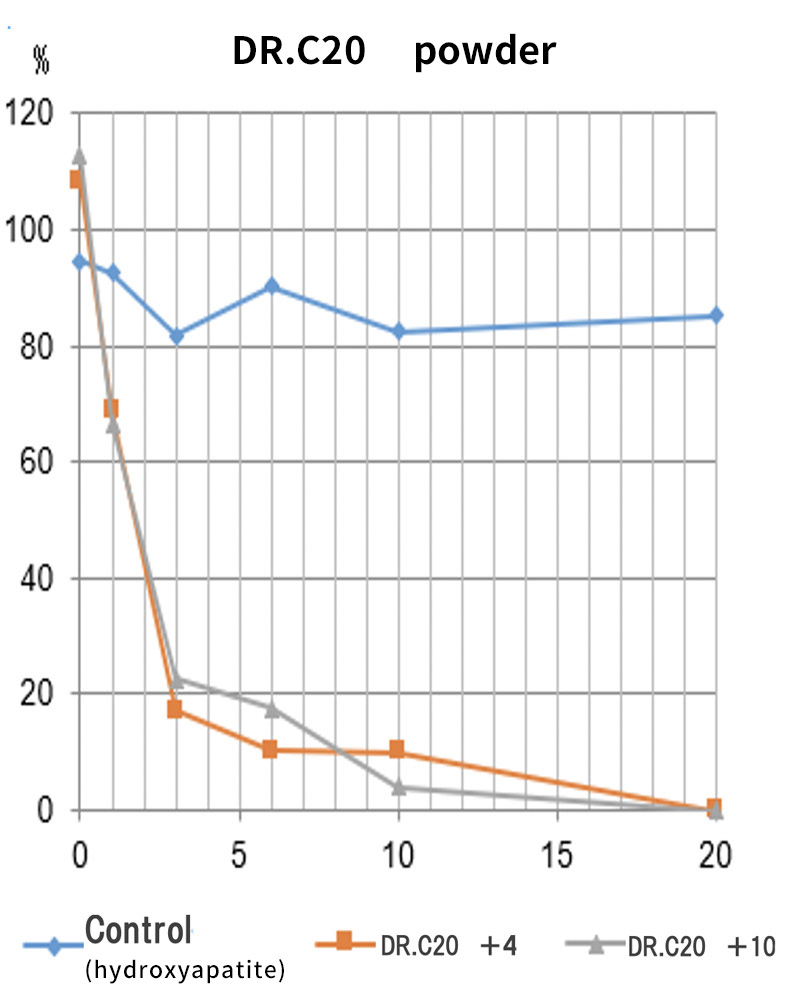
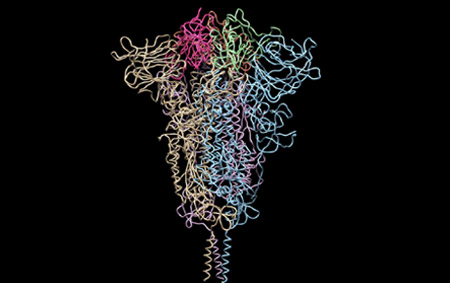
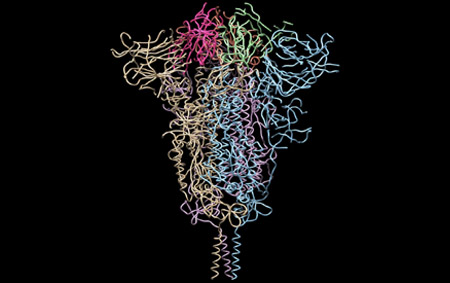
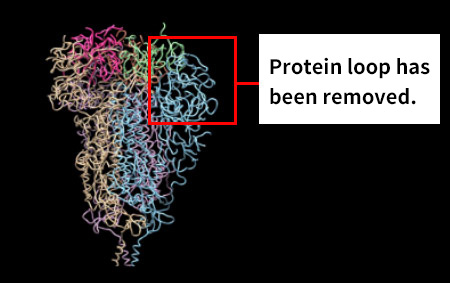
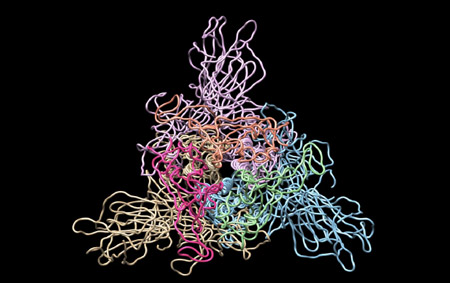
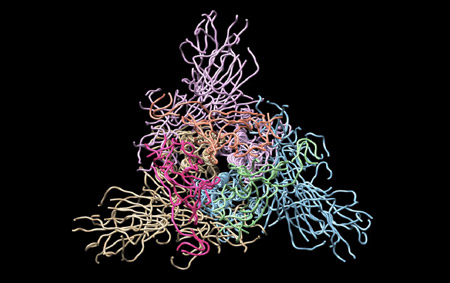
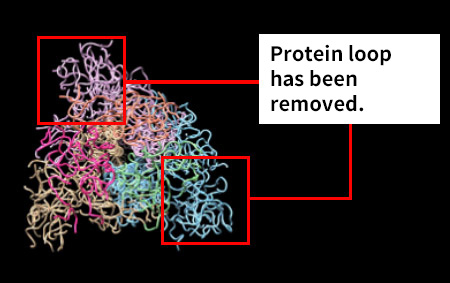
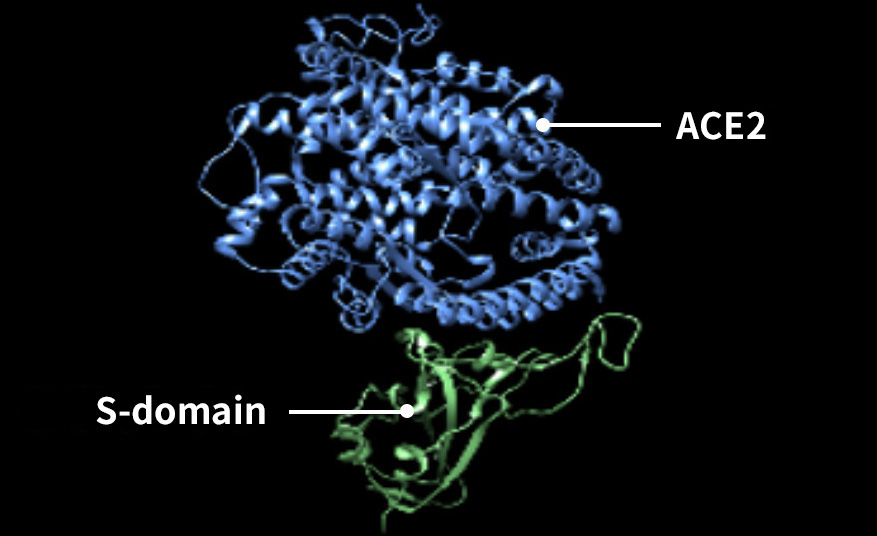
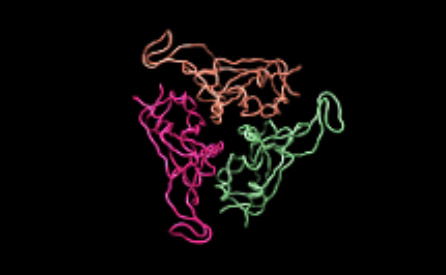
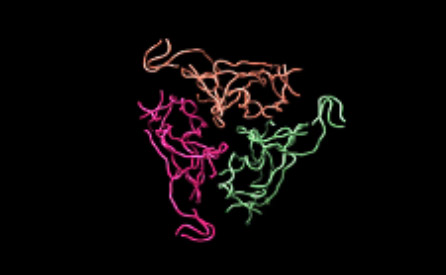
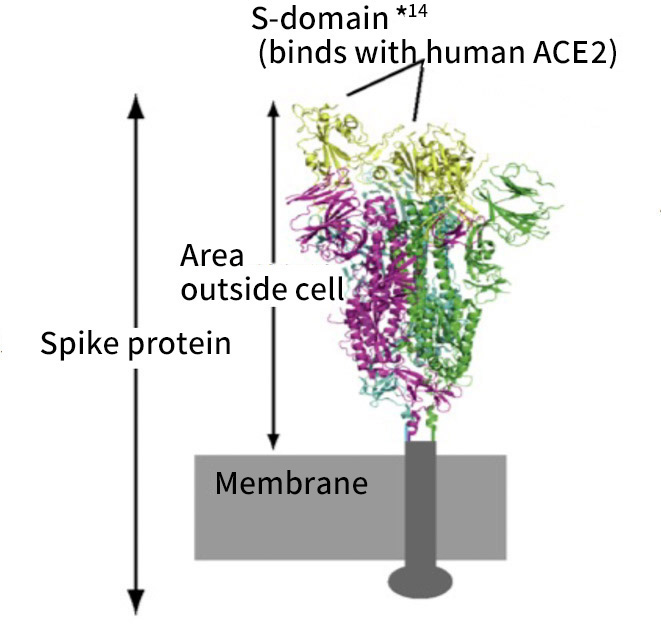
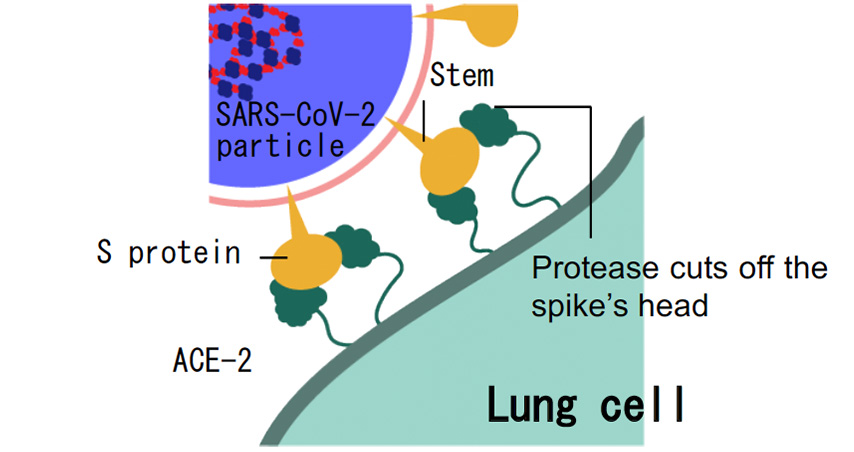
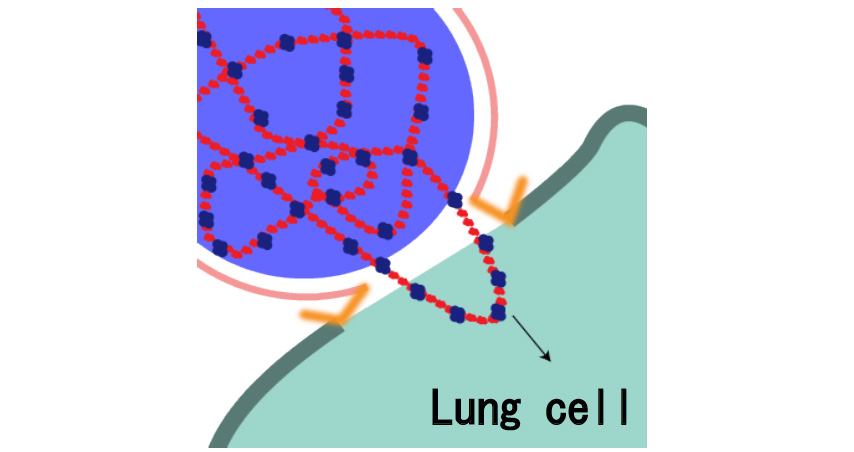
Moreover, together with Visiting Professor Shigeyuki Yokoyama of RIKEN’s Yokoyama Laboratory, an institution internationally renowned for protein synthesis, as a result of conducting an experiment using artificially-synthesized spike protein related to SARS-CoV-2 infection, we verified that DR.C20 cut the protein in multiple locations within a period of 3 to 10 minutes. The result of a molecular dynamics simulation carried out with the support of Associate Professor Toru Terada at the Graduate School of Agricultural and Life Sciences, The University of Tokyo, suggested that such cutting action by DR.C20 breaks down the 3D structure of the spike protein necessary for it to bond with the human cell receptor, ACE2. We clarified that these effects of DR.C20 on spike protein led to a reduction in the virus infectivity titer.